Biocomputing Graduate Project Series

Course: CSE 568 – Biocomputing
Institution: Arizona State University
Tools: Python, NumPy, Pandas, Seaborn, Matplotlib, Mesa (Agent-Based Modeling), Subprocess, Textor Algorithm
Institution: Arizona State University
Tools: Python, NumPy, Pandas, Seaborn, Matplotlib, Mesa (Agent-Based Modeling), Subprocess, Textor Algorithm
This graduate-level series explored computational biology through real-world problems in anomaly detection, agent-based modeling, and bio-inspired optimization. The projects applied Python programming and modeling frameworks to simulate biological behavior and analyze medical datasets.
Anomaly Detection in Cardiotocography: Implemented a negative selection algorithm to identify abnormal fetal heart rate and uterine contraction patterns. Achieved high AUC scores across varying detection thresholds using the Textor algorithm, applying string-based matching to binned medical data.
Agent-Based Modeling of Ant Behavior: Simulated collective foraging behavior using the Mesa library, capturing how simple local rules (discovery, recruitment, attrition) led to global decision-making among ant colonies. Validated results against ODE models to confirm behavioral trends.
Ant Colony Optimization (ACO) for Network Repair: Built an ACO algorithm to simulate pheromone-based path repair in broken network topologies. Tuned decay and exploration parameters to maximize route recovery success across minimal, simple, and medium graph structures.
Through these projects, I developed strong skills in modeling biological systems, detecting anomalies in health data, and applying bio-inspired algorithms to real-world optimization challenges—laying the groundwork for future research in computational healthcare.
Optimizing Glucose Level Predictions Through Data Clustering and Machine Learning



Course: CSE 572: Data Mining
Institution: Arizona State University
Tools: Python, NumPy, Pandas, Scikit-learn, SMOTE, KMeans, DBSCAN
Institution: Arizona State University
Tools: Python, NumPy, Pandas, Scikit-learn, SMOTE, KMeans, DBSCAN
This project focused on analyzing continuous glucose monitoring (CGM) and insulin pump data to develop predictive models and uncover patterns in diabetic patient behavior. It involved three components:
Time Series Analysis: Extracted 18 glucose control metrics (e.g., time in range, hyperglycemia) for manual and automated insulin delivery modes, segmented by daytime and overnight intervals.
Machine Learning Classification: Built a supervised learning model to detect meal vs. no-meal periods using statistical features (mean, std, FFT components) and balanced the dataset using SMOTE. Achieved moderate classification performance (F1 score: 0.613).
Unsupervised Clustering: Applied KMeans and DBSCAN to group meal data based on carbohydrate intake. Evaluated cluster quality using SSE, purity, and entropy, showing better results with KMeans.
Through this work, I gained hands-on experience in healthcare data preprocessing, feature extraction, model training, and validation, and I explored the real-world application of machine learning to improve diabetes management and support artificial pancreas systems.
Neural Network-based Robot Navigation/ A.I.
October 2023, Arizona State University
• Developed a neural network model for a simulated robotics task, aiming to help a small robot navigate its environment without collisions.
• Collected and managed a dataset, designed a custom neural network architecture using PyTorch.
Handwritten Digits Recognition / Machine Learning
September 2023, Arizona State University
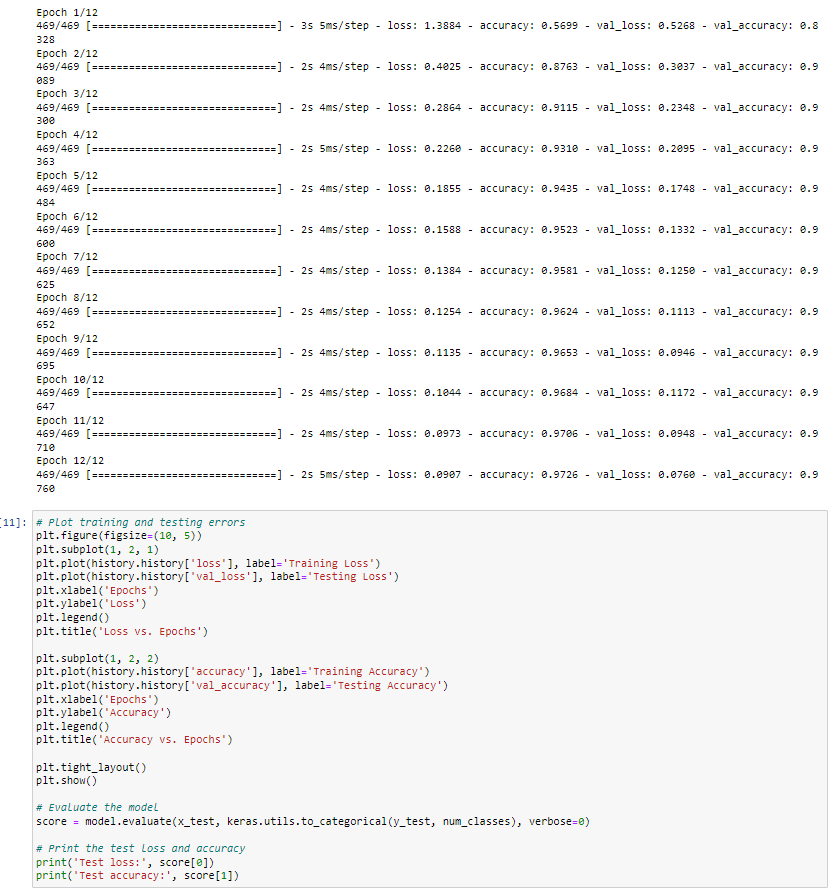
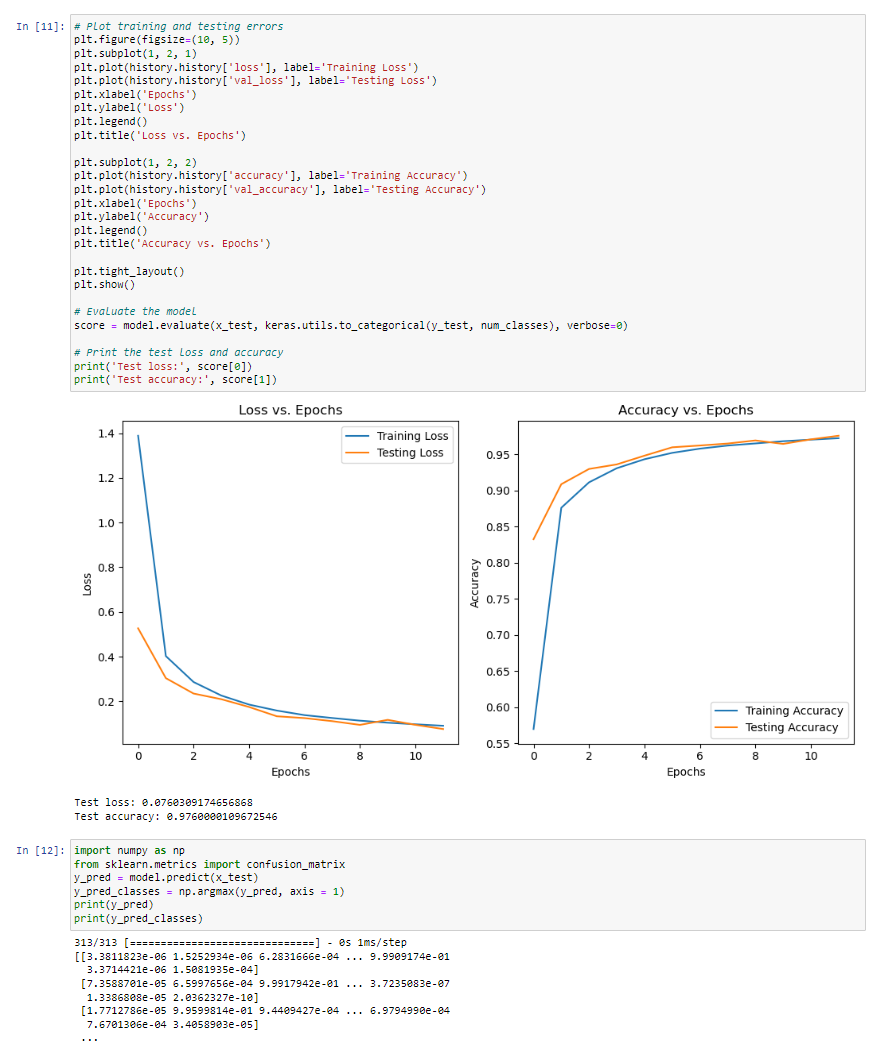
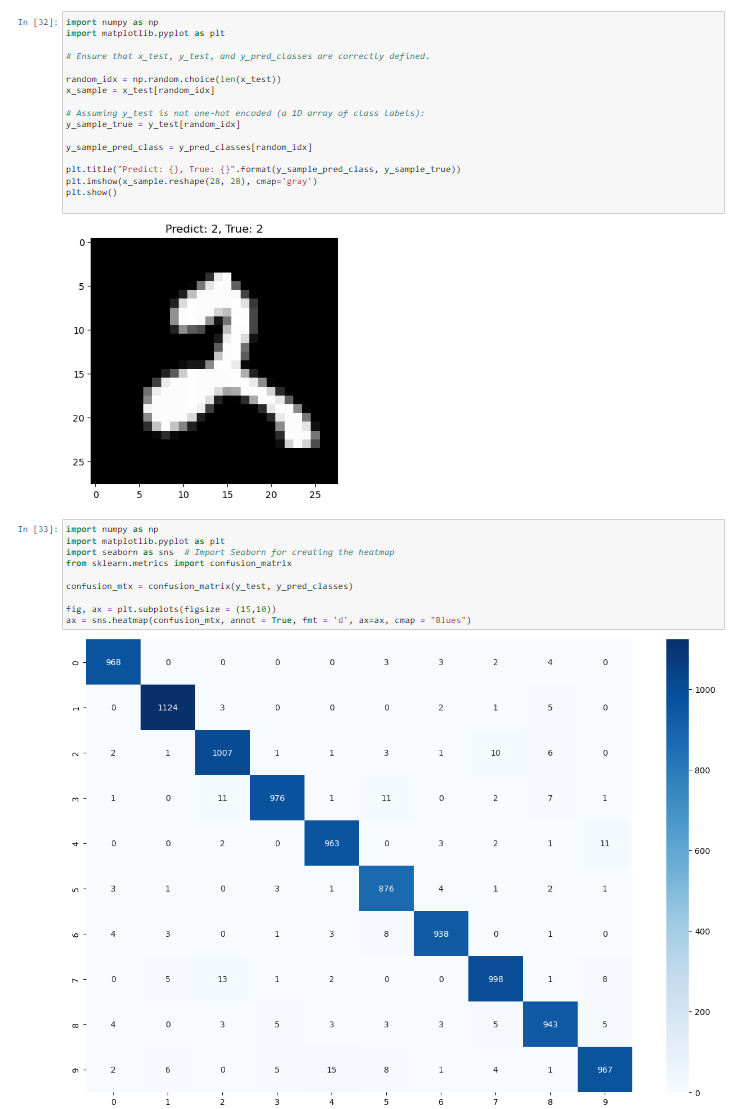
In this project, I implemented a machine learning model to recognize handwritten digits using the MNIST dataset, which contains 70,000 grayscale images of digits (0-9) labeled for classification. The dataset is widely used for benchmarking computer vision models. I preprocessed the images, applied feature extraction techniques, and trained a classification model using algorithms such as Convolutional Neural Networks (CNNs), Support Vector Machines (SVMs), and Random Forests. The goal was to optimize accuracy while minimizing computational cost. This project helped me gain hands-on experience in deep learning, computer vision, and hyperparameter tuning to improve model performance.
Snippets of part of the project code: